An effective direct-detection alternative for targeted ssRNAs
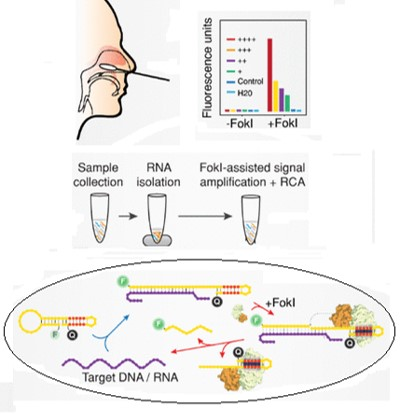
Representative scheme of the detection method
The design of new RNA direct detection approaches based on signal augmentation that avoids problems derived from nucleic acid amplification procedures in complex samples is still a challenge. Furthermore, there is a need since COVID 19 pandemic for alternatives that may prevent situations of collapse in healthcare systems.
Researchers from CSIC have validated a method where, after the specific attachment of the targeted material with a specially designed molecular beacon, the Fokl enzyme cleavages the DNA:RNA hybrid to release a fluorophore group, amplifying the signal exponentially due to the recirculation of the target sequence for further rounds of digestion.
This strategy reduces considerably the detection limits of targeted RNAs, demonstrating high specificity regardless of the complexity of the samples. It also presents multiplex capabilities, simultaneously detecting different sequences in the same sample. Moreover, it has been successfully coupled with isothermal amplification of nucleic acids techniques.
Main innovations and advantages
- The detection limit has been reduced more than 3 times in · RNA samples compared to conventional hybridization assays.
- The method provides specificity and multiplex capabilities, being able to discriminate between currently known · SARS-CoV-2 virus variants.
- It improves the detection limit of target molecules by 2 to 4 orders of magnitude when coupled with isothermal amplification of nucleic acids techniques.