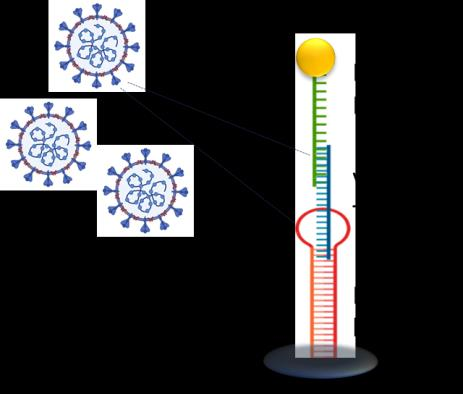
CSIC, the University of Barcelona and CIBER have developed novel DNA probes useful for the rapid detection of SARS-CoV2, influenza and other respiratory viral RNAs. Integrated in different set of devices, these probes enable the detection of RNA sequences in few minutes, without the need for RNA extraction, purification and PCR amplification.
Industrial partners from the diagnostic or pharmaceutical industry are being sought to collaborate through a patent licence agreement.
An offer for Patent Licensing
Rapid, inexpensive and sensitive device to detect viral infections
Results obtained from nasopharyngeal swabs of patients infected with SARS-CoV-2 by application of DNA probes in a thermal lateral flow device. Rapid and efficient testing is required to effectively control SARS-CoV-2 transmission. RT-qPCR is the current gold standard for population-scale testing. Although highly specific, its sensitivity in clinical practice is only 70% and requires long turnaround times (TAT) because of the need to extract the sample and amplify the viral RNA. Tests are performed on centralized laboratories, thus delaying even more the delivery of the tests results.
Other diagnostic technologies, such as antigen tests, can provide rapid results but present lower sensitivity and specificity.
Here is presented a fast, sensitive and reliable biochemical approach that allows performing the test at the point of care, without specialized equipment or highly trained personnel, or to use it on multiplexed high- throughput laboratory platforms to increase the efficiency and reduce TAT.
This method relies on the use of innovative DNA probes, based on polypurine reverse Hoogsteen hairpins (PPRHs), showing high affinity for RNA of SARS-CoV-2 and influenza viruses and forming triplex structures that present improved stability of the resulting complexes.
Four different diagnostic devices have been optimized (namely, thermal lateral flow PoC, an electrochemical PoC biosensor, ELISA microplate and fluorescent microarray) that are able to reach a detectability close to that achieved by other molecular methods, but without the need for PCR amplification.
Main innovations and advantages
· RNA virus quantification can be achieved in less than 20 min followingsample collection, without the need for RNA extraction, purificationand PCR amplification.
· High sensitivity: limits of detection comparable to those determined byRT-PCR in clinical samples obtained from nasopharyngeal swabs ofpatients infected with SARS-CoV-2 (up to 34 Ct, femtoM).
· Specificity: It has potential to differentiate SARS-CoV-2 from otherviruses such as influenza (H1N1).
· Useful for on-site routine diagnosis of SARS-CoV-2 and influenzaviruses as well as for high-throughput diagnostic screening.